You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
I have paired end fastq file from mgi sequencer using pcr free protocol. fastp --fix_mgi_id -w 16 --adapter_sequence AAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA --adapter_sequence_r2 AAGTCGGATCGTAGCCATGTCGTTCTGTGAGCCAAGGAGTTG -i 1.fq.gz -I 2.fq.gz -o fastp_1.fq.gz -O fastp_2.fq.gz -j fastp.json -h fastp.html
I explicitly mentioned adapters which are used in the experiment in the fastp command.
The result for adapters look like this, can you comment on what this indicates?
The text was updated successfully, but these errors were encountered:
I have paired end fastq file from mgi sequencer using pcr free protocol.
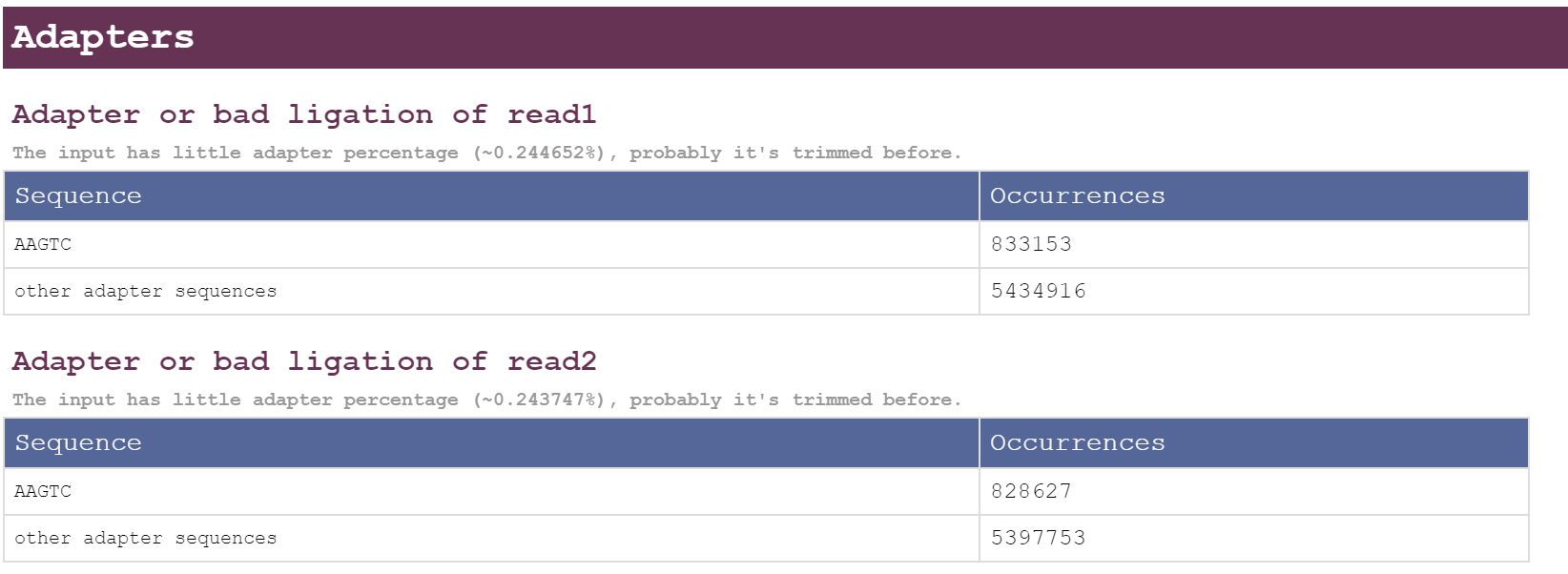
fastp --fix_mgi_id -w 16 --adapter_sequence AAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA --adapter_sequence_r2 AAGTCGGATCGTAGCCATGTCGTTCTGTGAGCCAAGGAGTTG -i 1.fq.gz -I 2.fq.gz -o fastp_1.fq.gz -O fastp_2.fq.gz -j fastp.json -h fastp.htmlI explicitly mentioned adapters which are used in the experiment in the fastp command.
The result for adapters look like this, can you comment on what this indicates?
The text was updated successfully, but these errors were encountered: